Harnessing Clinical Sequencing Data for Survival Stratification of Patients With Metastatic Lung Adenocarcinomas
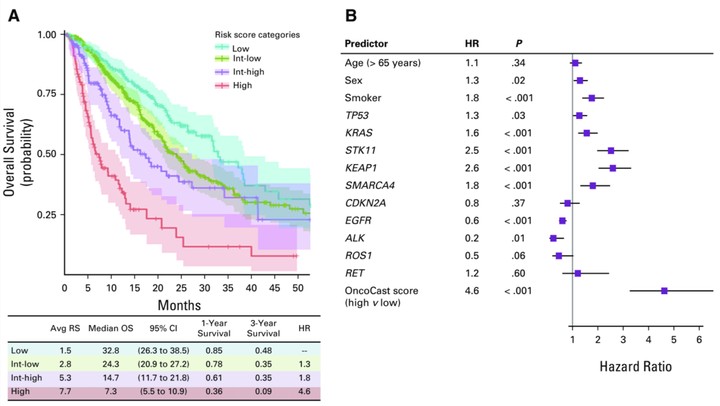 Image credit: ASCO Pubs
Image credit: ASCO Pubs
Abstract
Broad-panel sequencing of tumors facilitates routine care of people with cancer as well as clinical trial matching for novel genome-directed therapies. We sought to extend the use of broad-panel sequencing results to survival stratification and clinical outcome prediction. By using sequencing results from a cohort of 1,054 patients with advanced lung adenocarcinomas, we developed OncoCast, a machine learning tool for survival risk stratification and biomarker identification. With OncoCast, we stratified this patient cohort into four risk groups on the basis of tumor genomic profile. Patients whose tumors harbored a high-risk profile had a median survival of 7.3 months (95% CI, 5.5 to 10.9 months) compared with a low-risk group with a median survival of 32.8 months (95% CI, 26.3 to 38.5 months) with a hazard ratio of 4.6 (P < .001), far superior to any individual gene predictor or standard clinical characteristics. We found that comutations of both STK11 and KEAP1 are strong determinants of unfavorable prognosis with currently available therapies. In patients with targetable oncogenes (eg, EGFR, ALK, ROS1) who received targeted therapies, the tumor genetic background additionally differentiated survival with mutations in TP53 and ARID1A, which contributed to a higher risk score for shorter survival. A mutational profile derived from broad-panel sequencing presents an effective genomic stratification for patient survival in advanced lung adenocarcinoma. OncoCast is available as a public resource that facilitates the incorporation of mutational data to predict individual patient prognosis and compare risk characteristics of patient populations.